RANSAC example
In this example, we compare the performance of a normal algebraic one step (AOS) Pivot Calibration [Yaniv2010], with a RANSAC version of the same algorithm.
[1]:
%matplotlib inline
[2]:
# Jupyter notebook sets the cwd to the folder containing the notebook.
# So, you want to add the root of the project to the sys path, so modules load correctly.
import sys
sys.path.append("../../")
[3]:
# All imports.
from glob import glob
import copy
import random
import cv2
import numpy as np
import matplotlib.pyplot as plt
# Note that the scikit-surgery libraries provide pivot and RANSAC.
import sksurgerycalibration.algorithms.pivot as p # AOS Pivot algorithm and a RANSAC version.
import sksurgerycore.transforms.matrix as m # For creating 4x4 matrices.
[4]:
# Load test data
file_names = glob('PivotCalibrationData/*')
arrays = [np.loadtxt(f) for f in file_names]
matrices = np.concatenate(arrays)
number_of_matrices = int(matrices.size/16)
tracking_matrices = matrices.reshape(number_of_matrices, 4, 4)
print(tracking_matrices.shape)
(1151, 4, 4)
[7]:
# Do a normal pivot calibration using all data.
offset_1, pivot_1, residual_1 = p.pivot_calibration(tracking_matrices)
print("Offset is:" + str(offset_1))
print("Pivot is:" + str(pivot_1))
print("RMS error about centroid is:" + str(residual_1))
Offset is:[[-14.47617201]
[395.14282161]
[ -7.55790421]]
Pivot is:[[ -805.28473588]
[ -85.44779266]
[-2112.0664371 ]]
RMS error about centroid is:1.8384664420100074
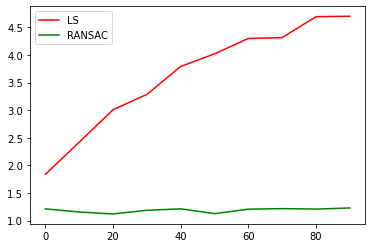
So, if we add noisy data, we should be able to see the difference between a normal pivot calibration (least squares), and the RANSAC version. We have about 1000 matrices, so lets add noise up to 100 of them.
[8]:
# Parameters to play with
number_of_matrices_to_add_noise_to = 100
rotation_offset_in_degrees = 5
translation_offset_in_millimetres = 5
number_iterations = 10
error_in_millimetres = 4
minimum_percentage = 0.25
[10]:
# Create an array representing how many to add noise to.
rms_error_ls = []
rms_error_RANSAC = []
x_values = []
indices = range(tracking_matrices.shape[0])
for counter in range(0, number_of_matrices_to_add_noise_to, 10):
matrices_copy = copy.deepcopy(tracking_matrices)
random_indices = random.sample(indices, counter)
for random_index in random_indices:
random_rotation = m.construct_rotm_from_euler((np.random.rand() * rotation_offset_in_degrees*2)-rotation_offset_in_degrees,
(np.random.rand() * rotation_offset_in_degrees*2)-rotation_offset_in_degrees,
(np.random.rand() * rotation_offset_in_degrees*2)-rotation_offset_in_degrees,
"zyx", is_in_radians=False)
random_translation = np.zeros((3,1))
random_translation[0][0] = (np.random.rand() * translation_offset_in_millimetres*2)-translation_offset_in_millimetres
random_translation[1][0] = (np.random.rand() * translation_offset_in_millimetres*2)-translation_offset_in_millimetres
random_translation[2][0] = (np.random.rand() * translation_offset_in_millimetres*2)-translation_offset_in_millimetres
random_transform = m.construct_rigid_transformation(random_rotation, random_translation)
matrices_copy[random_index] = matrices_copy[random_index] @ random_transform
offset_2, pivot_2, residual_2 = p.pivot_calibration(matrices_copy)
offset_3, pivot_3, residual_3 = p.pivot_calibration_with_ransac(matrices_copy, number_iterations, error_in_millimetres, minimum_percentage)
x_values.append(counter)
rms_error_ls.append(residual_2)
rms_error_RANSAC.append(residual_3)
RANSAC Pivot, from 1151 matrices, used 1010 matrices, with error threshold = 4 and consensus threshold = 0.25
RANSAC Pivot, from 1151 matrices, used 924 matrices, with error threshold = 4 and consensus threshold = 0.25
RANSAC Pivot, from 1151 matrices, used 925 matrices, with error threshold = 4 and consensus threshold = 0.25
RANSAC Pivot, from 1151 matrices, used 966 matrices, with error threshold = 4 and consensus threshold = 0.25
RANSAC Pivot, from 1151 matrices, used 952 matrices, with error threshold = 4 and consensus threshold = 0.25
RANSAC Pivot, from 1151 matrices, used 899 matrices, with error threshold = 4 and consensus threshold = 0.25
RANSAC Pivot, from 1151 matrices, used 897 matrices, with error threshold = 4 and consensus threshold = 0.25
RANSAC Pivot, from 1151 matrices, used 920 matrices, with error threshold = 4 and consensus threshold = 0.25
RANSAC Pivot, from 1151 matrices, used 911 matrices, with error threshold = 4 and consensus threshold = 0.25
RANSAC Pivot, from 1151 matrices, used 948 matrices, with error threshold = 4 and consensus threshold = 0.25
[11]:
plt.plot(x_values, rms_error_ls, 'r', label='LS')
plt.plot(x_values, rms_error_RANSAC, 'g', label='RANSAC')
plt.legend(loc='upper left')
plt.show()
[ ]: